Hallmarks of Biological Failure
Category: Application Area Application Area: Biological Failure
Date/Time: April 8, 2019 - April 10, 2019
Group photo
 (x)
(x)
Organizers
Michael Hochberg (Univ. Montpellier/SFI)
Daniel Promislow (Univ. Washington)
Bernie Crespi (Simon Fraser Univ.) - Complexity, Breaking Bad Tradeoffs, and the Evolution of Biological Failure[edit source]
I will discuss two points. First, how are genomic and organismal complexity related to slow, and fast, failure? Do biological systems fail more under conditions of high
complexity and tight coupling, as posited for inanimate systems? Do increases in genomic and organismal complexity result in short-term benefits, but more longer-term evolutionary vulnerabilities? Second, how do tradeoffs mediate failure? Most tradeoffs are 'bad' in that system-wide organismal lifetime optimization is not achieved, even if they are relatively 'good' for propagating genes. Can such bad tradeoffs be broken, artificially, by humans?
I think so, in some cases. I discuss examples, from mental disorders, life histories, and senescence.
Barbara Natterson-Horowitz (Harvard) - Dynamic Cardiovascular Systems, Evolved Adaptations and Clinical Pathology[edit source]
Several forms of high impact human cardiovascular(CV) pathology are related to autonomic dysregulation, emerging in association with adrenergic events. A phylogenetic survey of organisms with spontaneous occurrence of these pathologies and correlation with life history points to the adaptive value of the phenotypic flexibility facilitated by these dynamic systems. It also suggests that varied intra-individual CV physiologic responses to environmental threat are complex, adaptive and play a central role in vulnerability to several forms of human cardiovascular pathology.
James DeGregori (CU Denver) - Explain this! - Evolutionary approaches to unanswered questions in cancer biology[edit source]
I plan to throw out some observations (e.g. age-dependent cancer incidence for different organs and different species), and how these are currently enigmatic. I’ll discuss possible explanations, but also highlight were explanations are currently lacking.
David Schneider (Stanford) - Measuring the resilience of hosts to infections by mapping disease space[edit source]
My group has been trying to find relatively simple multidimensional ways of measuring the response to infections. Our idea is to measure how far a host will be pushed from its normal physiology when it sickens and what route it will take coming back from sickness. We do this by drawing the trajectory infected individuals take through phase space and try to produce maps that improve our understanding of the process. We want to understand how far the system can be pushed before it breaks, which is one sort of system failure. We then want to understand how this varies. For example, do hosts die because their physiology becomes more elastic? In this case they would be more likely to enter physiological states that are not survivable. Alternatively, physiological states that would be survivable when to one host might not be survivable to another. Our first project is to understand what variation looks like when we examine infections this way. As we proceed we would like to model this system more carefully.
Rozalyn Anderson (Univ. Wisconsin) - Metabolic Integrity & Aging: Amplification of Small Perturbations[edit source]
Caloric restriction (CR) delays aging and the onset of age-related disease in diverse species, including nonhuman primates. Emerging data has focused our studies on links between metabolic status and disease vulnerability; several diseases of aging including diabetes, cancer, and neurodegeneration, have an established metabolic component. Candidate factors involved in longevity regulation are nutrient sensitive and interconnected in terms of signaling pathways and downstream effector actions. Molecular profiling of the transcriptome, proteome, and metabolome identifies CR responsive elements that are highly enriched for metabolic pathways. Here too connectivity among responsive nodes, or mega clusters, is complex. Our recent work shows that small changes in metabolic status precipitate large-scale multi-modal functional changes across diverse cellular processes. We suggest that modest failures in metabolic integrity are amplified by such mechanisms with age to broadly impact homeostasis and adaptation, creating shared vulnerability to diseases and conditions despite differences in their etiology.
Shripad Tuljapurkar (Stanford) - Models in Aging: Two Examples[edit source]
I discuss approaches to two problems on very different timescales. For a single lifetime, transitions between states of health (disability) can be viewed as stochastic movement out of a potential with two minima. Aging can mean changes in the amplitude of noise, depth of potential, or width of potential. Such dynamics are conceptually similar to the disability transition in current medical understanding. What are the math features? Can we make this into a statistical model? On evolutionary timescales, post-reproductive life can evolve according to varipus arguments that are all examples of “borrowed fitness.” I explain what this means and mainly ask what questions we should be asking.
Marten Scheffer (Wageningen Univ.) - Quantifying Resilience of Humans and other Animals[edit source]
All life requires the capacity to recover from challenges that are as inevitable as they are unpredictable. Understanding this resilience is essential for managing the health of humans and their livestock. It has long been difficult to quantify resilience directly, forcing practitioners to rely on indirect static indicators of health. However, measurements from wearable electronics and other sources now allow us to analyze the dynamics of physiology and behavior with unsurpassed resolution. The resulting flood of data coincides with the emergence of novel analytical tools for estimating resilience from the pattern of micro-recoveries observed in natural time series. Such dynamic indicators of resilience (DIORs) may be used to monitor the risk of systemic failure across systems ranging from organs to entire organisms. These tools invite a fundamental rethink of our approach to the adaptive management of health and resilience.
Dario Riccardo Valenzano (Max Planck) - Relaxed selection shapes the rate of aging across species[edit source]
African killifishes independently evolved annual life cycles at least three times, offering a unique natural experiment of diversification of life history strategies. Using a comprehensive whole-genome sampling of 46 species of African killifishes, we found that genome size correlates with annual life style and climate. Annual species had genome-wide expansion of transposable elements, higher gene family turn-over rates and relaxed selection in genes in known aging pathways, such as mitochondrial replication and translation, mTOR pathway and DNA repair. Whole-genome resequencing in wild Nothobranchius populations showed bottle-necks and a genome-wide signature of relaxation of selection in populations evolved in dryer climates. In conclusion, evolution in ephemeral environments in African killifishes caused an extensive relaxation of selective constraints at genome-wide level. We discovered that, in African killifishes, ecology drove the evolution of short life span and rapid aging, associated to tens of thousands of slightly deleterious mutations driven to high frequencies.
Sabrina Spencer (CU Boulder) - Single-cell analysis of heterogeneity in proliferation-quiescence decisions[edit source]
Research in the Spencer lab is focused on understanding how signaling events control cell fate. Studying these processes in single cells reveals remarkable cell-to-cell variability in response to stimuli, even among genetically identical cells in a uniform environment. We seek to understand the sources and consequences of this heterogeneity in the cellular response to stimuli. The stimuli we study include growth factors, cell stress, and targeted cancer therapeutics. To do this, we develop genetically encoded fluorescent sensors for signaling events of interest. We then use long-term live-cell microscopy and cell tracking to quantify the dynamics of upstream signals and link them to cell fate (proliferation, quiescence, apoptosis, senescence). Our long-term goal is to understand the normal mechanistic functioning of signaling pathways that control proliferation, to understand how these signals go awry in cancer, and eventually to alter the fate of individual cells.
Morgan Levine (Yale Univ.) - Systems-Level Modeling of Aging across Biological Levels of Organization[edit source]
Aging is associated with numerous changes at all levels of biological organization. Harnessing this information to develop measures that accurately and reliably quantify the biological aging process will require incorporation of functioning/failure at various levels that can be integrated using systems level approaches. This talk will provide illustrations on how DNA methylation data (DNAm) can be integrated with cellular, physiological, proteomic, and clinical data to model age-related changes that propagate up the levels—finally manifesting as age-related disease or death. We will also show how network modeling can be used to generate a ‘diseasome’ model in order to identify hub methylation signatures with implication for multiple pathways and outcomes. Given the complexity of the biological aging process, modeling of systems dynamics over time will both lead to the development of better biomarkers of aging, and also inform our conceptualization of how alterations at the molecular level propagate up levels of organization to eventually influence morbidity and mortality risk.
Daniel Promislow (Univ. Washington) Link to the source page[edit source]
April 9, 2019
Charge for working groups
- Come up with a list of major ideas/problems/concepts that you think we need to work on.
- Think about what conceptual areas could be linked to better address the major questions discussed in #1.
Apr 8, 2019
A few thoughts about general questions for discussion:
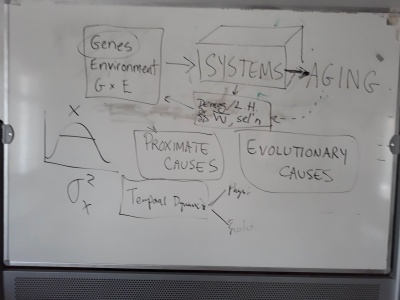
- What do we mean by "Biological Failure"? Aging? Senescence?
- Is there such a thing as a truly non-aging organism? An immortal organism?
- Things that change with age...
- Why do so many things appear to increase exponentially, and in parallel on a log-linear scale, with age?
- Are there commonalities across other levels of organization with respect to how things change with age, and by 'things', this could be function, or selection, or failure.
- What maintains variance among populations in aging. Even after controlling for G and E, we still see high levels of variance.
- Issues of complexity/simplicity and networks were touched upon today, but we have not yet gone into detail, discussing this.
Bernie Crespi
Suggestions idea that complexity can be bad. We can think of organismal organization (and function and failure?) in a two-dimensional space of couple (loose<-->tight) and complexity (linear<-->high). Berni suggests (I think) that biological entities with high complexity (brains, immune system) are more prone to bring the entire system down with failure. Entities that are simple and loosely coupled are less likely to fail badly. Suggests a negative correlation between senescence and intelligence, though the GWAS data supporting this are problematic (like the recent study claiming to find genes for SES--https://www.biorxiv.org/content/10.1101/457515v1) are likely due to social stratification. Bernie's neologism of the day: badaptation.
Dario Valenzano
Comparative analysis of annual life history in killifish. Would benefit from Nathan Clark approach to look at rates of protein evolution in annual/perennial species: https://elifesciences.org/articles/25884. Expansion of mitochondrial genome is striking. I wonder if the finding of increase in genome size in the annual species is true of annual plant species (like corn and rice, which have very large genomes) relative to perennial plants.
James deGregori
Notion of adaptive oncogenesis, with stem cells well adapted to niche. As tissue ages, stem cells are no longer well adapted to the niche in which they find themselves. I wonder whether these models are also relevant to the selection process that happens *within* a tumor once cancer growth (and mutator phenotypes) is underway.
Roz Anderson
CR animals show a few major clusters of correlated -omic features, while AL animals show a very large number of small molecules. We should discuss just what these correlations mean, both statistically and biologically, and why these correlation structures (adjacency matrices) differ among the two groups so dramatically.
Tulja
Finishes talk with three general questions:
a. How strong is the trade-off between added longevity and lost fertility
b. Can we explain environmental plasticity?
c. Does stochasticity matter? On any time scale?
For all three of these questions, I wonder if high-dimensional assays (metabolome, epigenome, etc) might have something to add to this discussion...
Sabrina Spencer
The technology that Sabrina is development could add tremendous power to the work now ongoing to track yeast cells as they age in real time. Also shows that quiescent cells are resistant to various stressors. Is that simply that they are metabolically quiescent and so not taking in these toxins?
Barb Natterson-Horowitz
Suggests that the disease-associated heart responses that we see could be maladaptive responses that evolved for adaptive reasons ('capture myopathy', 'alarm bradycardia'). Barb finds evidence for these phenomena in non-human species, but these could well be a large underestimate, simply because the vast majority of these events are never observed, and when observed, not reported.
Michael Hochberg (Univ. Montpellier/SFI) Link to the source page[edit source]
Reference Materials by Presenting Attendees[edit source]